Generates either a variability decomposition plot of factor
effects or a diagnostic plot for an object of class eda_npol.
Usage
# S3 method for class 'eda_npol'
plot(x, plot = "effects", reg = FALSE, ...)Arguments
- x
An object of class
eda_npol.- plot
A character string specifying the type of plot to generate.
"effects"(default)Generates a variability decomposition plot showing residuals and factor effects.
"diagnostic"Generates a scatterplot of residuals versus comparison values (CV).
- reg
Logical. If
TRUE, fits a linear regression line to the diagnostic plot. Only used whenplot = "diagnostic". Defaults toFALSE.- ...
Additional arguments passed to internal plotting functions.
- For
plot = "effects" Arguments are passed to
.eda_plot_vardecomp. Common options include:rotate: Logical. Rotate plot orientation.show.resp: Logical. Include boxplot of the centered response.outliers: Logical. Show outliers in boxplots.label: Logical. Label individual effect levels.order: Logical. Order effects by spread.cex.txt: Numeric. Text size for labels.lim: Numeric vector. Axis limits.overlap: Character. One of"stack","overplot", or"jitter".grey: Numeric or character. Grayscale coloring.type: Character. Plot type, e.g.,"boxpnt"or"box".input: Character. Either"nway"or"reg".padding: Numeric. Padding for axis limits.
- For
plot = "diagnostic" Arguments are passed to
.eda_plot_xy. Common options include:xlab,ylab: Axis labels.xlim,ylim: Axis limits.poly: Integer. Degree of polynomial regression.robust: Logical. Use robust regression.w: Numeric vector. Weights for regression.sd,mean.l: Logical. Show ±1 SD and mean lines.asp,square: Logical. Control aspect ratio and plot shape.grey: Numeric. Grayscale background.pch,p.col,p.fill,size,alpha: Point styling.q,inner,q.type,qcol: Quantile box options.loe,loe.col,loe.lw,loess.d: Loess smoothing options.lm.col,lm.lw: Regression line styling.stats,stat.size: Display model statistics.hline,vline: Reference lines.rlm.d: List. Parameters forMASS::rlm.
- For
Details
This method serves as a wrapper to generate two types of plots for
eda_npol objects:
1. Variability Decomposition Plot (plot = "effects")
Calls .eda_plot_vardecomp to visualize residuals and factor
effects. Useful for assessing the relative magnitude and spread of effects
and identifying outliers.
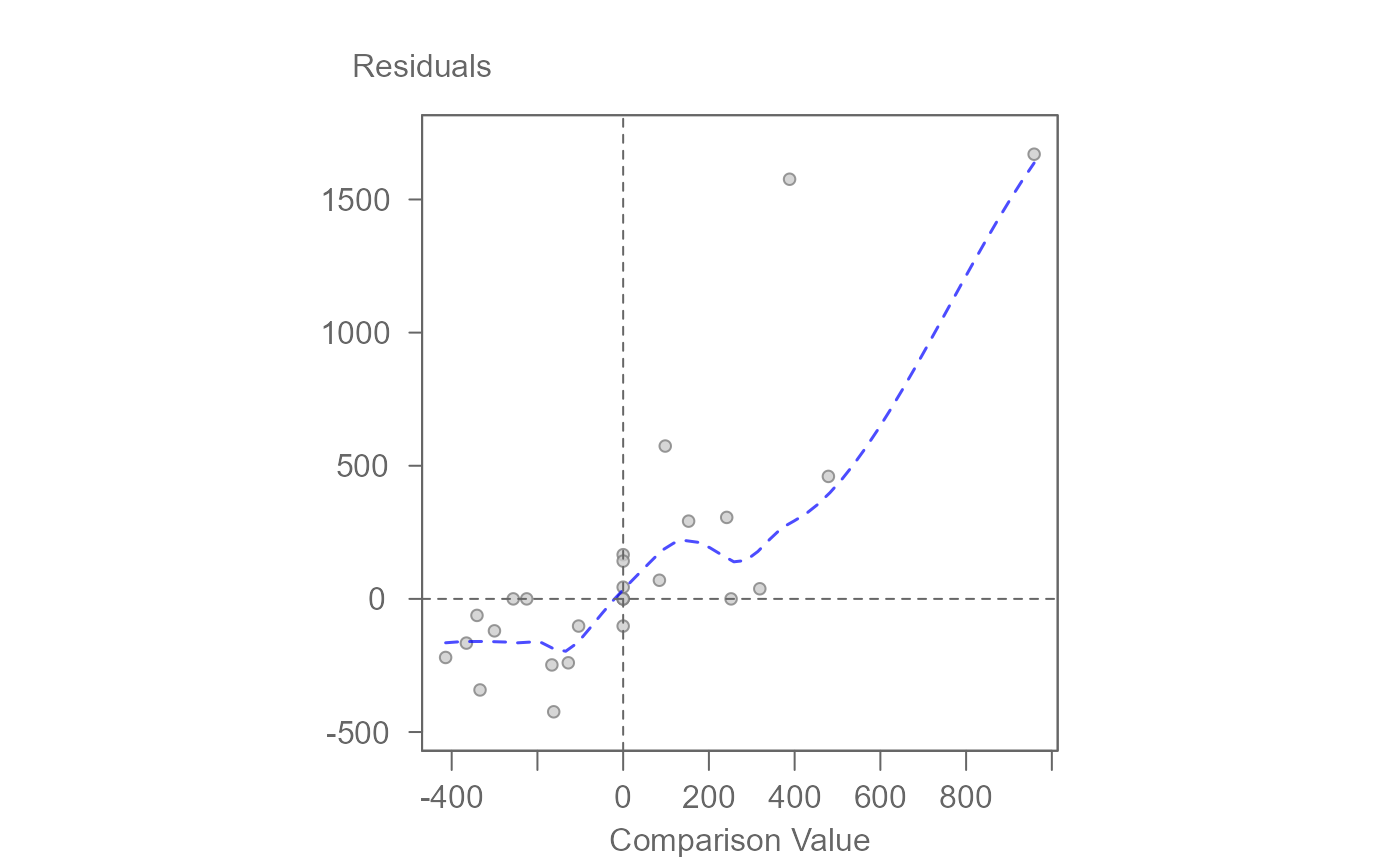
2. Diagnostic Plot (plot = "diagnostic")
Calls
.eda_plot_xy to plot residuals against comparison values
(CV). Useful for detecting nonadditivity or model misfit. Optional
regression and smoothing lines can be added.
Examples
# Generate eda_npol object
M0 <- eda_npol(yarn, Cycles, Load, Length, Amplitude)
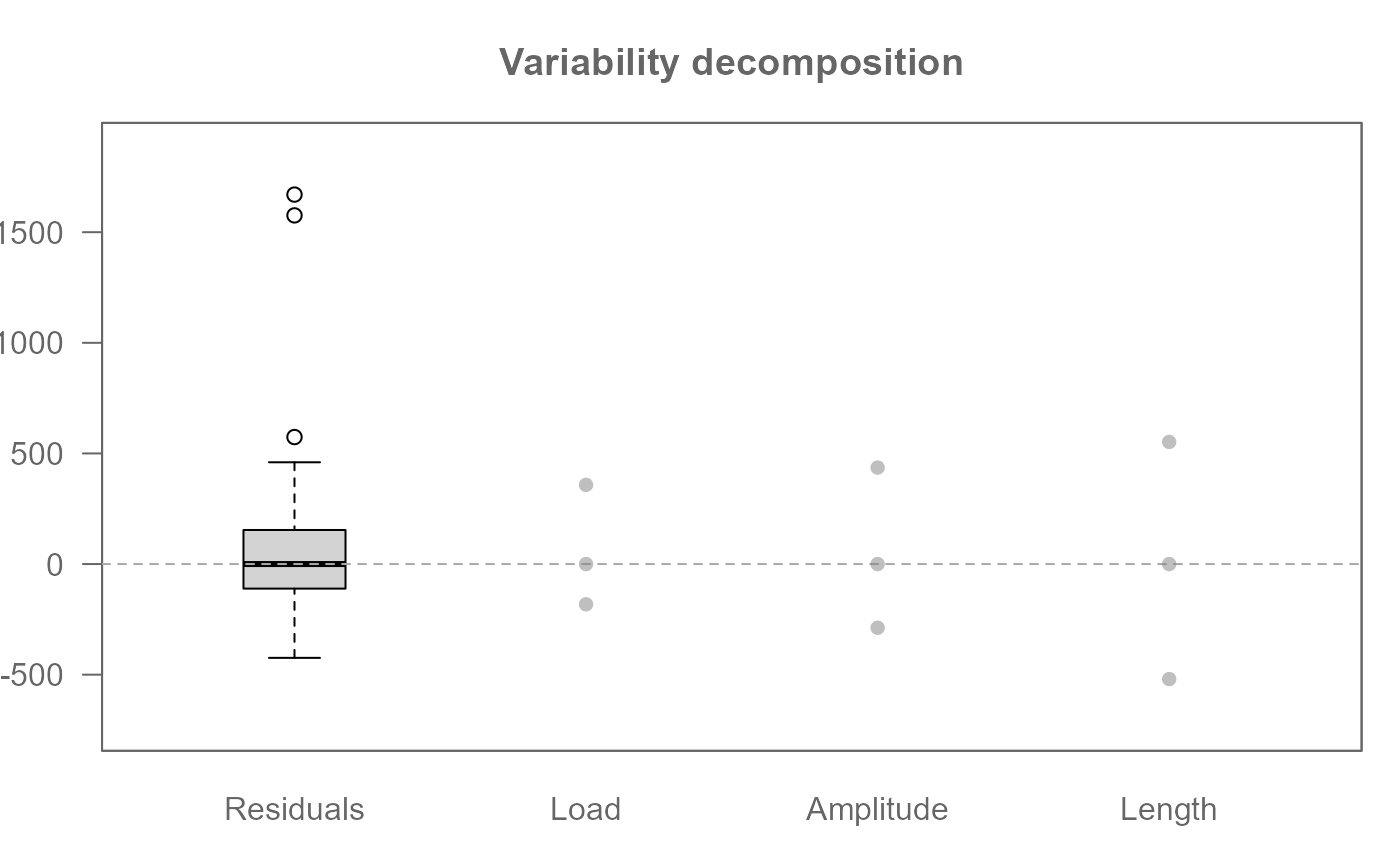
# Plot effects (default)
plot(M0)
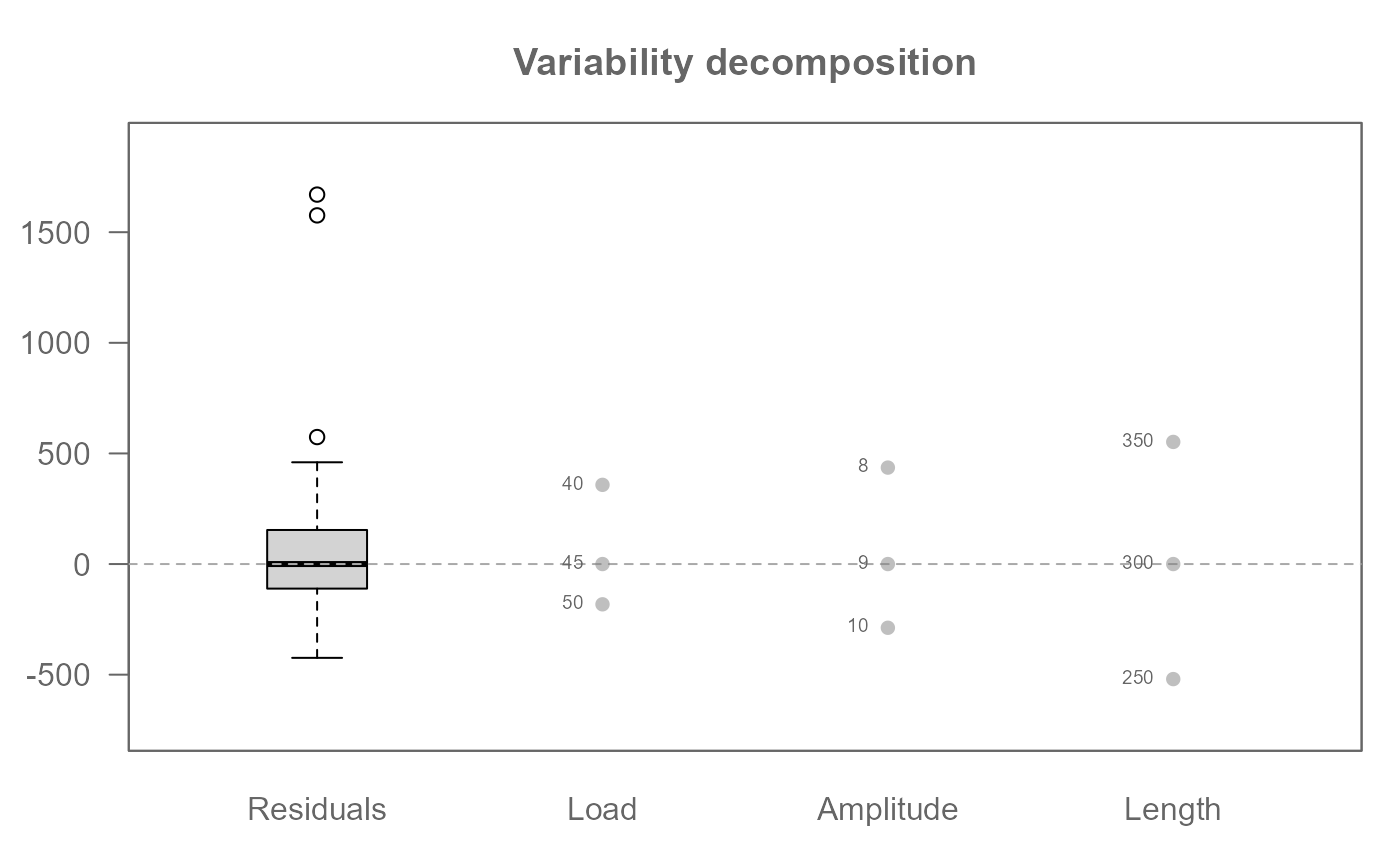
 # Add labels
plot(M0, label = TRUE)
# Add labels
plot(M0, label = TRUE)
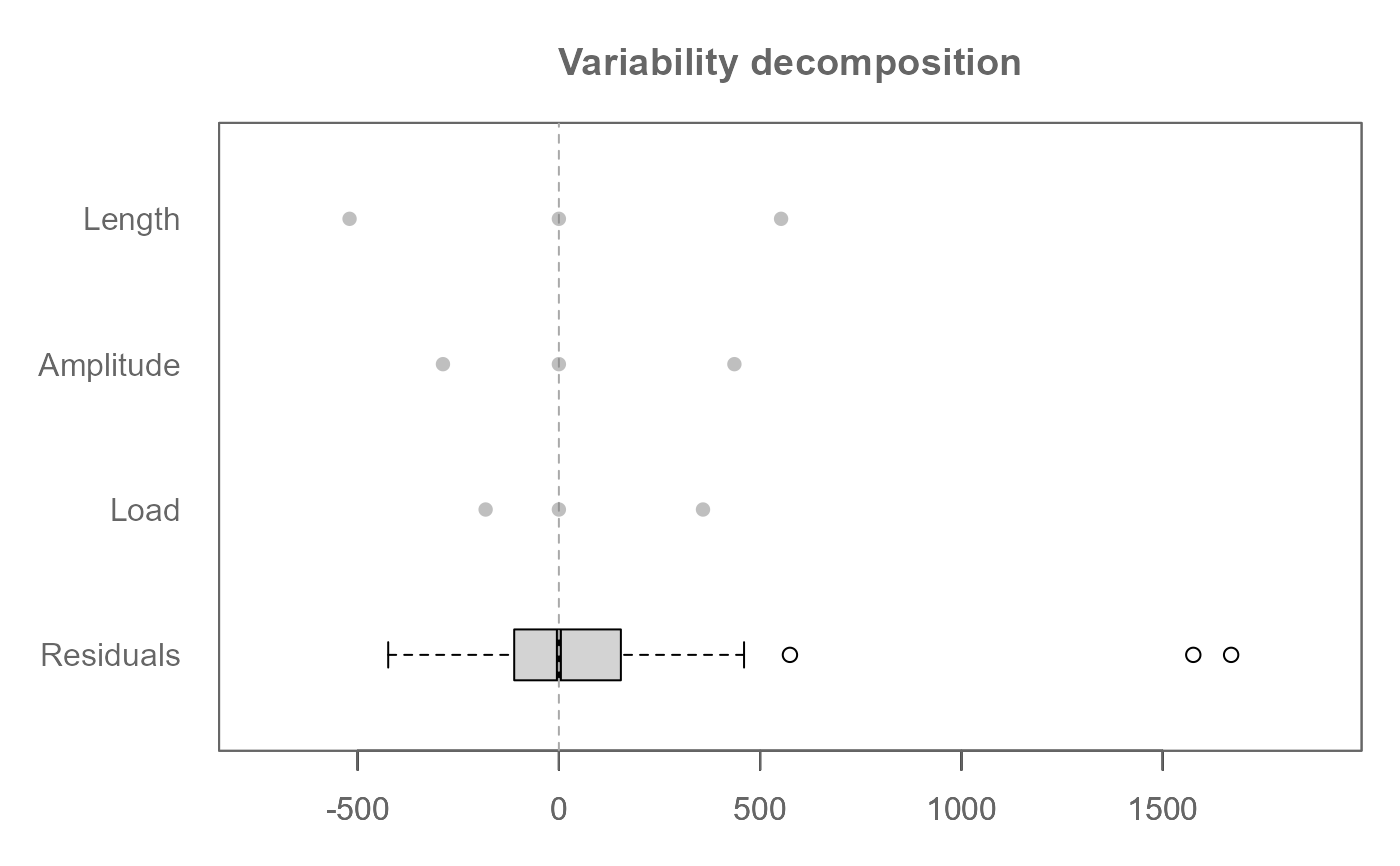
 # Rotate plot
plot(M0, rotate = TRUE)
# Rotate plot
plot(M0, rotate = TRUE)
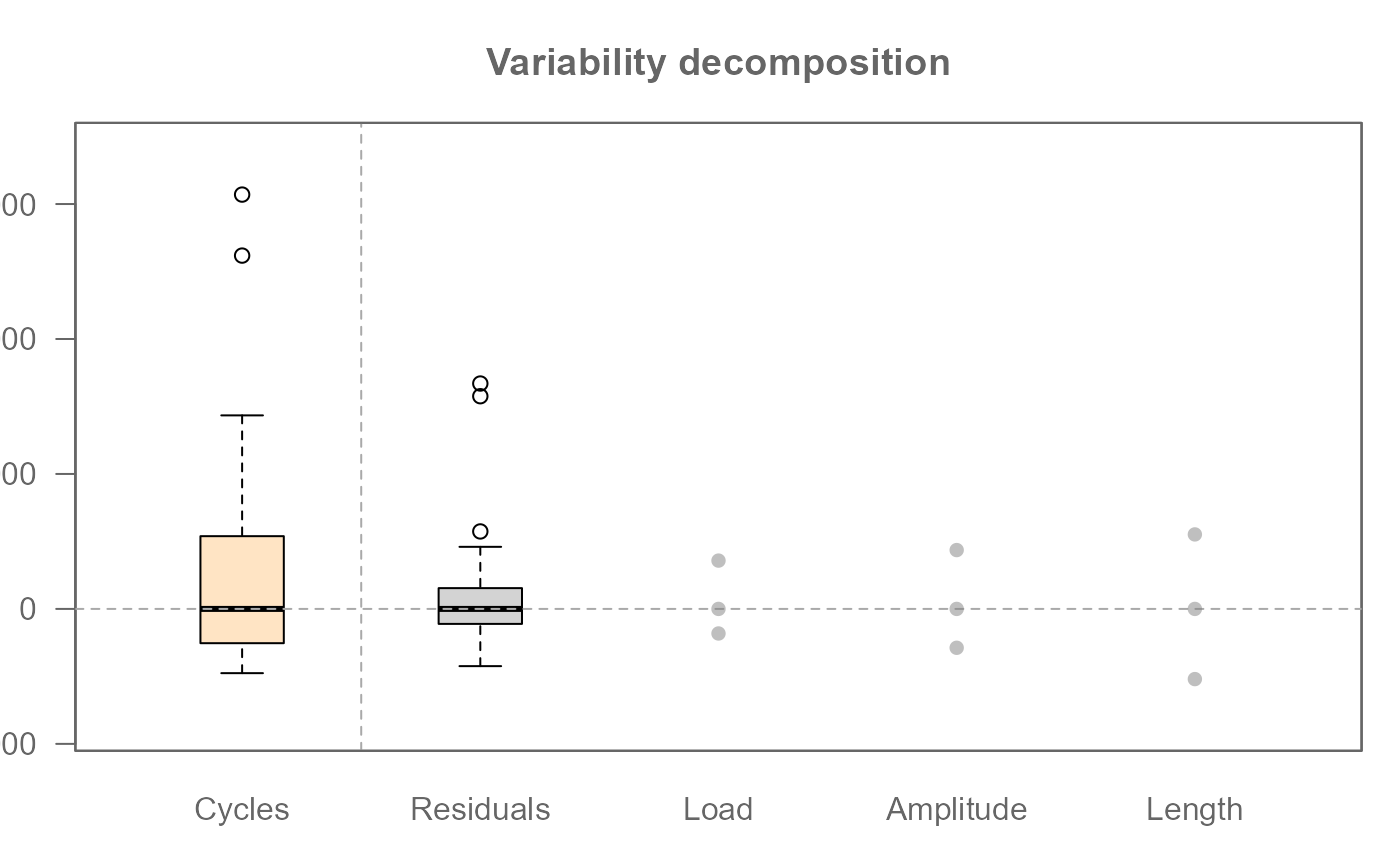
 # Add boxplot of centered response variable
plot(M0, show.resp = TRUE)
# Add boxplot of centered response variable
plot(M0, show.resp = TRUE)
 # Generate diagnostic plot (residuals vs CV)
plot(M0, plot = "diagnostic")
# Generate diagnostic plot (residuals vs CV)
plot(M0, plot = "diagnostic")
 # Fit a robust regression line to diagnostic plot
# The function displays the line's slope in the console
plot(M0, plot = "diagnostic", reg = TRUE, robust = TRUE, loe = FALSE)
# Fit a robust regression line to diagnostic plot
# The function displays the line's slope in the console
plot(M0, plot = "diagnostic", reg = TRUE, robust = TRUE, loe = FALSE)
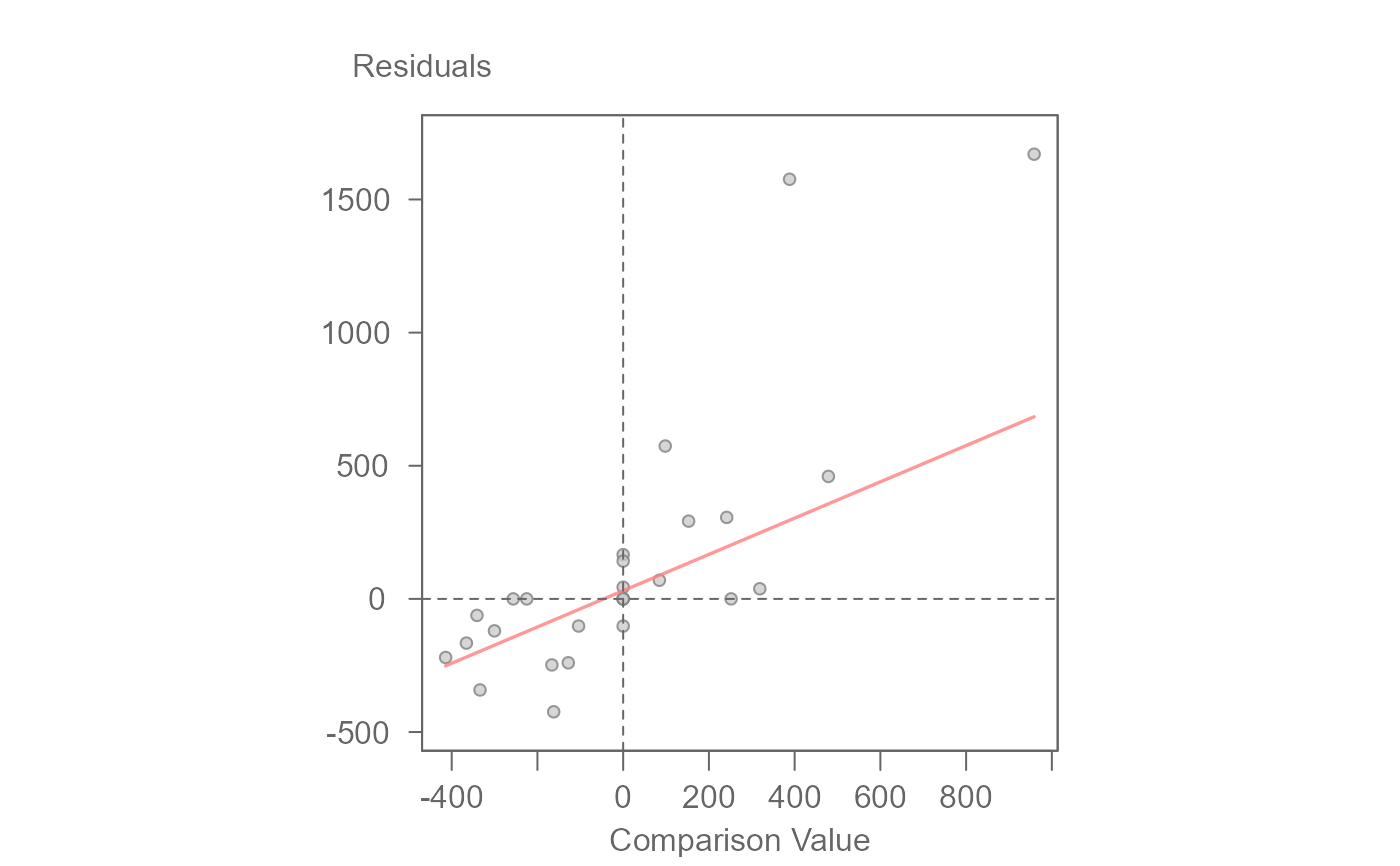 #> int Comparison Value^1
#> 30.7565973 0.6813374
#> int Comparison Value^1
#> 30.7565973 0.6813374